RavenCAD: a DNA assebly planning tool
Published:
RavenCAD is a dynamic programing algorithm for planing DNA synthesis protocols that is accessible as a web application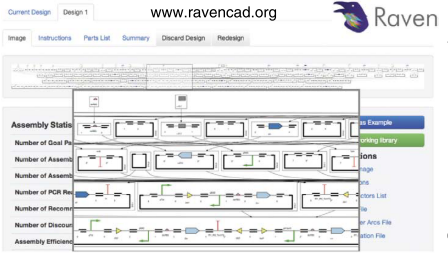
Published:
RavenCAD is a dynamic programing algorithm for planing DNA synthesis protocols that is accessible as a web application
Published:
TBA is a multi-functional machine learning tool for identifying words in the genome (aka transcription factor motifs) associated with genomic features.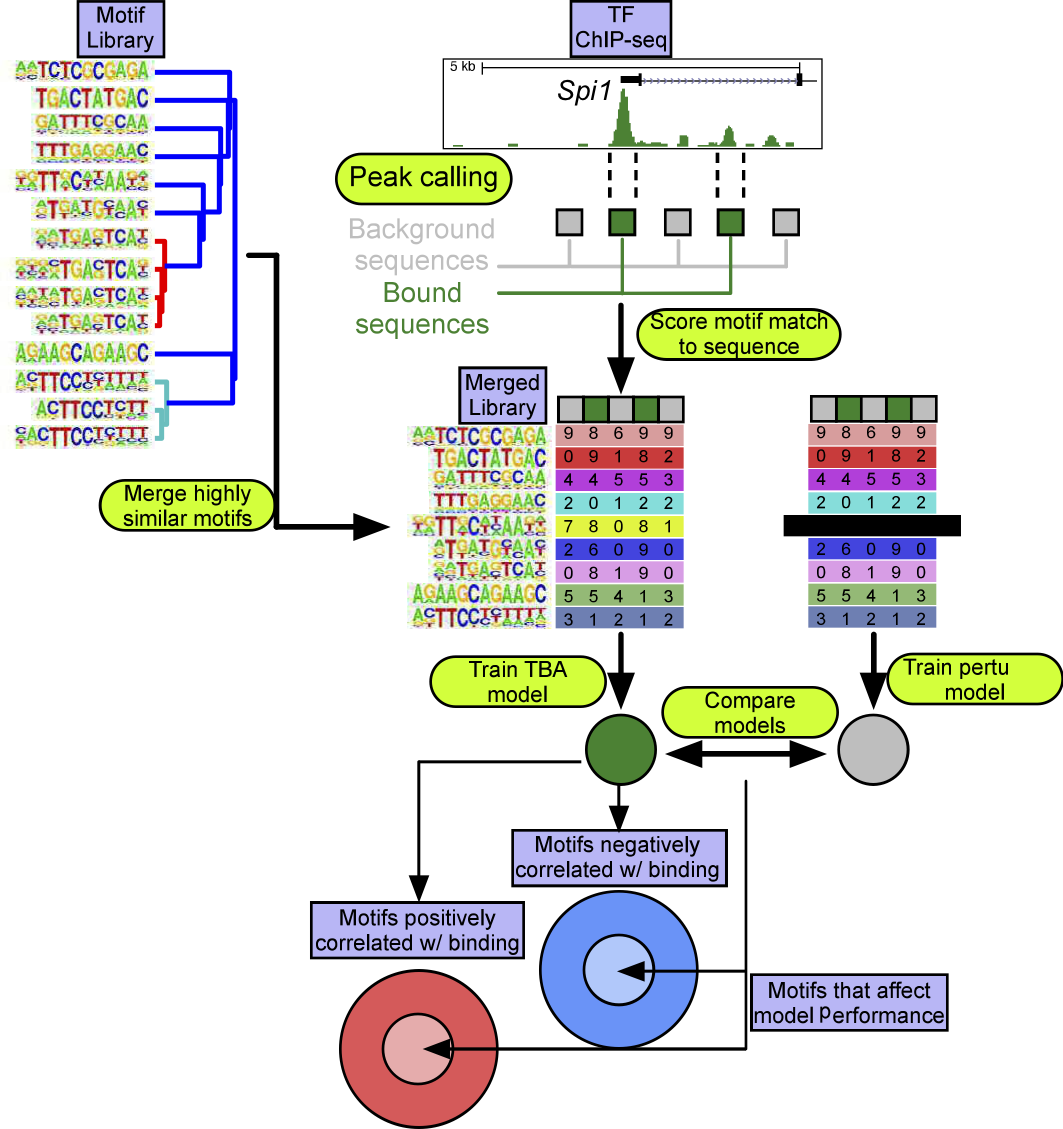
Published:
A series of neural networks and algorithms for identifying a genomic grammar that can be used to classify regulatory circuits in the mammalian genome. The current model, is a convolutional network that uses a dot-product attention mechanism. This network has performance on par with or exceeding current state-of-the-art methods such as DeepBind at distinguishing regulatory circuits from random genomic background. Additionally, the attention mechanism of the model can be used to extract the architectures of regulatory circuits in the genome via a post-processing step performed on the output of the model.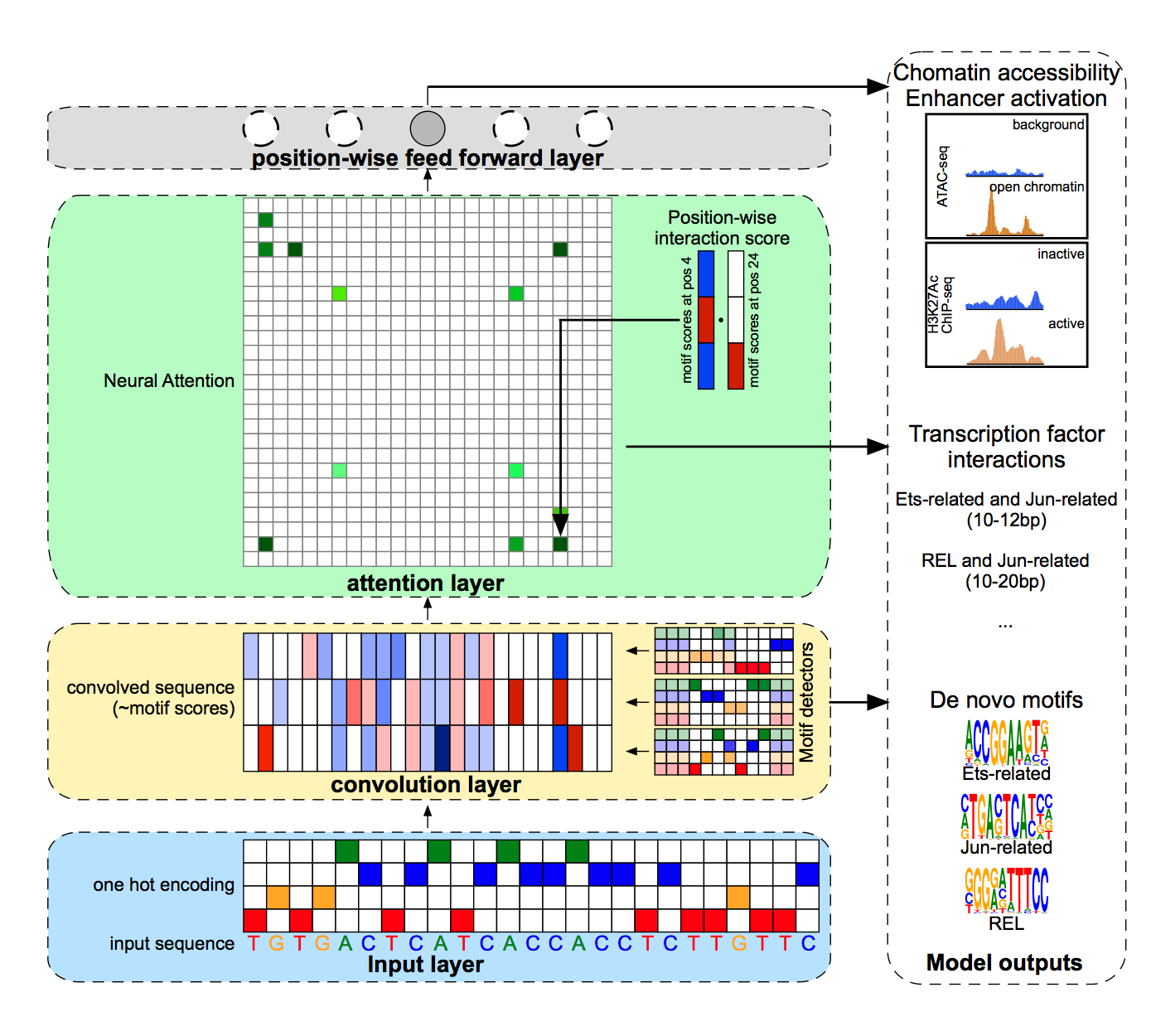
Published in Nature Methods, 2014
This paper describes RavenCAD, a dynamic programing algorithm for planing DNA synthesis protocols that is accessible as a web application.
Download here
Published in ACS Synthetic Biology, 2014
This paper describes RavenCAD, a dynamic programing algorithm for planing DNA synthesis protocols that is accessible as a web application.
Download here
Published in eLife, 2016
This paper investigates transcription factors that regulate the wound-healing response using genomics techniques
Download here
Published in Cell Metabolism, 2017
In this paper, the authors identify a biphasic macrophage gene expression program underlying anti-inflammatory fatty acid production following TLR4 activation. The late anti-inflammatory program is dependent on SREBP1. This is surprising given the known roles of SREBP1 in promoting IL1B production during the induction of inflammation and highlights its dual roles.
Download here
Published in Proceedings of the National Academy of Sciences, 2018
This paper describes novel cell-specific differences in LXR responses to natural and synthetic ligands in macrophages and liver cells that provide a conceptually new basis for future drug development.
Download here
Published in Cell, 2018
Analyses of genetic variation in macrophages from inbred mouse strains reveal how a complex network of transcription factors influence cis- regulatory elements to impact differentiation and responses to environmental change.
Download here
Published in BioRxiv, 2018
This paper describes the application of a logistic regression model that uses a feature set, which is programmatically curated to reduce multiple collinearity, to identify combinations of sequence features targeted by transcription factors within the AP-1 transcription factor family.
Download here
Published in Scientific Reports, 2018
This study examines a gene expression signature in the circulating monocytes of Parkinson’s disease patients that can be used as a potential biomarker early in the disease course.
Download here
Published in International Workshop on Bio-Design Automation, 2018
This paper describes the application of a convolutional nerual network with an attention mechanism that can identify regulatory circuits in the genome with state-of-the-art performance. The attention mechanism of the neural network can also reveal the architecture of regulatory circuits within the genome
Download here
Published in AAAI Fall Symposium: Artificial Intelligence for Synthetic Biology, 2018
This paper describes the application of a neural attention mechanism for studying the architecture of regulatory elements within the genome
Download here
Published in University 1, Department, 2013
This is a description of your workshop, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published in University 1, Department, 2014
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published in University 1, Department, 2014
This is a conference proceedings talk, for when you give a talk at a conference that archives talks as publications. When the talk_type variable is set to “Conference proceedings talk”, this talk will appear in the Talks page in /talks/, but it will not appear in the Talks section in the /cv/ page. The reason for this is that the conference proceedings talk should have a seprate publications .md file in the /_publications/ folder, which will go in the publications section. You can change this behavior by modifying the /_pages/cv.md file.
Published in Test University, Department of Testing Science, 2016
This talk is an introduction to Testing.
Published in University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Published in University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.